Pandas (continues)¶
[1]:
import pandas as pd
import numpy as np
Catenating datasets¶
We already saw in the NumPy section how we can catenate arrays along an axis: axis=0 catenates vertically and axis=1 catenates horizontally, and so on. With the DataFrames of Pandas it works similarly except that the row indices and the column names require extra attention. Also note a slight difference in the name: np.concatenate but pd.concat.
Let’s start by considering catenation along the axis 0, that is, vertical catenation. We will first make a helper function to easily create DataFrames for testing.
[2]:
def makedf(cols, ind):
data = {c : [str(c) + str(i) for i in ind] for c in cols}
return pd.DataFrame(data, ind)
Next we will create some example DataFrames:
[3]:
a=makedf("AB", [0,1])
a
[3]:
| A | B | |
|---|---|---|
| 0 | A0 | B0 |
| 1 | A1 | B1 |
[4]:
b=makedf("AB", [2,3])
b
[4]:
| A | B | |
|---|---|---|
| 2 | A2 | B2 |
| 3 | A3 | B3 |
[5]:
c=makedf("CD", [0,1])
c
[5]:
| C | D | |
|---|---|---|
| 0 | C0 | D0 |
| 1 | C1 | D1 |
[6]:
d=makedf("BC", [2,3])
d
[6]:
| B | C | |
|---|---|---|
| 2 | B2 | C2 |
| 3 | B3 | C3 |
In the following simple case, the concat function works exactly as we expect it would:
[7]:
pd.concat([a,b]) # The default axis is 0
[7]:
| A | B | |
|---|---|---|
| 0 | A0 | B0 |
| 1 | A1 | B1 |
| 2 | A2 | B2 |
| 3 | A3 | B3 |
The next, however, will create duplicate indices:
[8]:
r=pd.concat([a,a])
r
[8]:
| A | B | |
|---|---|---|
| 0 | A0 | B0 |
| 1 | A1 | B1 |
| 0 | A0 | B0 |
| 1 | A1 | B1 |
[9]:
r.loc[0,"A"]
[9]:
0 A0
0 A0
Name: A, dtype: object
This is not usually what we want! There are three solutions to this. Firstly, deny creation of duplicated indices by giving the verify_integrity parameter to the concat function:
[10]:
try:
pd.concat([a,a], verify_integrity=True)
except ValueError as e:
import sys
print(e, file=sys.stderr)
Indexes have overlapping values: Int64Index([0, 1], dtype='int64')
Secondly, we can ask for automatic renumbering of rows:
[11]:
pd.concat([a,a], ignore_index=True)
[11]:
| A | B | |
|---|---|---|
| 0 | A0 | B0 |
| 1 | A1 | B1 |
| 2 | A0 | B0 |
| 3 | A1 | B1 |
Thirdly, we can ask for hierarchical indexing. The indices can contain multiple levels, but on this course we don’t consider hierarchical indices in detail. Hierarchical indices can make a two dimensional array to work like higher dimensional array.
[12]:
r2=pd.concat([a,a], keys=['first', 'second'])
r2
[12]:
| A | B | ||
|---|---|---|---|
| first | 0 | A0 | B0 |
| 1 | A1 | B1 | |
| second | 0 | A0 | B0 |
| 1 | A1 | B1 |
[13]:
r2["A"]["first"][0]
[13]:
'A0'
Everything works similarly, when we want to catenate horizontally:
[14]:
pd.concat([a,c], axis=1)
[14]:
| A | B | C | D | |
|---|---|---|---|---|
| 0 | A0 | B0 | C0 | D0 |
| 1 | A1 | B1 | C1 | D1 |
We have so far assumed that when concatenating vertically the columns of both DataFrames are the same, and when joining horizontally the indices are the same. This is, however, not required:
[15]:
pd.concat([a,d], sort=False) # sort option is used to silence a deprecation message
[15]:
| A | B | C | |
|---|---|---|---|
| 0 | A0 | B0 | NaN |
| 1 | A1 | B1 | NaN |
| 2 | NaN | B2 | C2 |
| 3 | NaN | B3 | C3 |
It expanded the non-existing cases with NaNs. This method is called an outer join, which forms the union of columns in the two DataFrames. The alternative is inner join, which forms the intersection of columns:
[16]:
pd.concat([a,d], join="inner")
[16]:
| B | |
|---|---|
| 0 | B0 |
| 1 | B1 |
| 2 | B2 |
| 3 | B3 |
Write function split_date_continues that does
- read the bicycle data set
- clean the data set of columns/rows that contain only missing values
- drops the
Päivämääräcolumn and replaces it with its splitted components as before
Use the concat function to do this. The function should return a DataFrame with 25 columns (first five related to the date and then the rest 20 conserning the measument location.
Merging dataframes¶
Merging combines two DataFrames based on some common field.
Let’s recall the earlier DataFrame about wages and ages of persons:
[17]:
df = pd.DataFrame([[1000, "Jack", 21], [1500, "John", 29]], columns=["Wage", "Name", "Age"])
df
[17]:
| Wage | Name | Age | |
|---|---|---|---|
| 0 | 1000 | Jack | 21 |
| 1 | 1500 | John | 29 |
Now, create a new DataFrame with the occupations of persons:
[18]:
df2 = pd.DataFrame({"Name" : ["John", "Jack"], "Occupation": ["Plumber", "Carpenter"]})
df2
[18]:
| Name | Occupation | |
|---|---|---|
| 0 | John | Plumber |
| 1 | Jack | Carpenter |
The following function call will merge the two DataFrames on their common field, and, importantly, will keep the indices aligned. What this means is that even though the names are listed in different order in the two frames, the merge will still give correct result.
[19]:
pd.merge(df, df2)
[19]:
| Wage | Name | Age | Occupation | |
|---|---|---|---|---|
| 0 | 1000 | Jack | 21 | Carpenter |
| 1 | 1500 | John | 29 | Plumber |
This was an example of a simple one-to-one merge, where the keys in the Name columns had 1-to-1 correspondence. Sometimes not all the keys appear in both DataFrames:
[20]:
df3 = pd.concat([df2, pd.DataFrame({ "Name" : ["James"], "Occupation":["Painter"]})], ignore_index=True)
df3
[20]:
| Name | Occupation | |
|---|---|---|
| 0 | John | Plumber |
| 1 | Jack | Carpenter |
| 2 | James | Painter |
[21]:
pd.merge(df, df3) # By default an inner join is computed
[21]:
| Wage | Name | Age | Occupation | |
|---|---|---|---|---|
| 0 | 1000 | Jack | 21 | Carpenter |
| 1 | 1500 | John | 29 | Plumber |
[22]:
pd.merge(df, df3, how="outer") # Outer join
[22]:
| Wage | Name | Age | Occupation | |
|---|---|---|---|---|
| 0 | 1000.0 | Jack | 21.0 | Carpenter |
| 1 | 1500.0 | John | 29.0 | Plumber |
| 2 | NaN | James | NaN | Painter |
Also, many-to-one and many-to-many relationships can occur in merges:
[23]:
books = pd.DataFrame({"Title" : ["War and Peace", "Good Omens", "Good Omens"] ,
"Author" : ["Tolstoi", "Terry Pratchett", "Neil Gaiman"]})
books
[23]:
| Title | Author | |
|---|---|---|
| 0 | War and Peace | Tolstoi |
| 1 | Good Omens | Terry Pratchett |
| 2 | Good Omens | Neil Gaiman |
[24]:
collections = pd.DataFrame([["Oodi", "War and Peace"],
["Oodi", "Good Omens"],
["Pasila", "Good Omens"],
["Kallio", "War and Peace"]], columns=["Library", "Title"])
collections
[24]:
| Library | Title | |
|---|---|---|
| 0 | Oodi | War and Peace |
| 1 | Oodi | Good Omens |
| 2 | Pasila | Good Omens |
| 3 | Kallio | War and Peace |
All combinations with matching keys (Title) are created:
[25]:
libraries_with_books_by = pd.merge(books, collections)
libraries_with_books_by
[25]:
| Title | Author | Library | |
|---|---|---|---|
| 0 | War and Peace | Tolstoi | Oodi |
| 1 | War and Peace | Tolstoi | Kallio |
| 2 | Good Omens | Terry Pratchett | Oodi |
| 3 | Good Omens | Terry Pratchett | Pasila |
| 4 | Good Omens | Neil Gaiman | Oodi |
| 5 | Good Omens | Neil Gaiman | Pasila |
Merge the processed cycling data set (from the previous exercise) and weather data set along the columns year, month, and day. Note that the names of these columns might be different in the two tables: use the left_on and right_on parameters. Then drop useless columns ‘m’, ‘d’, ‘Time’, and ‘Time zone’.
Write function cycling_weather that reads the data sets and returns the resulting DataFrame.
Merge the DataFrames UK top40 and the bands DataFrame that are stored in the src folder. Do all this in the parameterless function top_bands, which should return the merged DataFrame. Use the left_on and right_on parameters to merge. Test your function from the main function.
Aggregates and groupings¶
Let us use again the weather dataset. First, we make the column names a bit more uniform and concise. For example the columns Year, m, and d are not uniformly named.
We can easily change the column names with the rename method of the DataFrame. Note that we cannot directly change the index wh.columns as it is immutable.
[26]:
wh = pd.read_csv("https://www.cs.helsinki.fi/u/jttoivon/dap/data/fmi/kumpula-weather-2017.csv")
[27]:
wh3 = wh.rename(columns={"m": "Month", "d": "Day", "Precipitation amount (mm)" : "Precipitation",
"Snow depth (cm)" : "Snow", "Air temperature (degC)" : "Temperature"})
wh3.head()
[27]:
| Year | Month | Day | Time | Time zone | Precipitation | Snow | Temperature | |
|---|---|---|---|---|---|---|---|---|
| 0 | 2017 | 1 | 1 | 00:00 | UTC | -1.0 | -1.0 | 0.6 |
| 1 | 2017 | 1 | 2 | 00:00 | UTC | 4.4 | -1.0 | -3.9 |
| 2 | 2017 | 1 | 3 | 00:00 | UTC | 6.6 | 7.0 | -6.5 |
| 3 | 2017 | 1 | 4 | 00:00 | UTC | -1.0 | 13.0 | -12.8 |
| 4 | 2017 | 1 | 5 | 00:00 | UTC | -1.0 | 10.0 | -17.8 |
Pandas has an operation that splits a DataFrame into groups, performs some operation on each of the groups, and then combines the result from each group into a resulting DataFrame. This split-apply-combine functionality is really flexible and powerful operation. In Pandas you start by calling the groupby method, which splits the DataFrame into groups. In the following example the rows that contain measurements from the same month belong to the same group:
[28]:
groups = wh3.groupby("Month")
groups
[28]:
<pandas.core.groupby.groupby.DataFrameGroupBy object at 0x7f9714311320>
Nothing happened yet, but the groupby object knows how the division into groups is done. This is called a lazy operation. We can query the number of groups in the groupby object:
[29]:
len(groups)
[29]:
12
We can iterate through all the groups:
[30]:
for key, group in groups:
print(key, len(group))
1 31
2 28
3 31
4 30
5 31
6 30
7 31
8 31
9 30
10 31
11 30
12 31
[31]:
groups.get_group(2) # Group with index two is February
[31]:
| Year | Month | Day | Time | Time zone | Precipitation | Snow | Temperature | |
|---|---|---|---|---|---|---|---|---|
| 31 | 2017 | 2 | 1 | 00:00 | UTC | 1.5 | 4.0 | -0.6 |
| 32 | 2017 | 2 | 2 | 00:00 | UTC | 0.2 | 5.0 | -0.8 |
| 33 | 2017 | 2 | 3 | 00:00 | UTC | -1.0 | 6.0 | -0.2 |
| 34 | 2017 | 2 | 4 | 00:00 | UTC | 2.7 | 6.0 | 0.4 |
| 35 | 2017 | 2 | 5 | 00:00 | UTC | -1.0 | 7.0 | -2.5 |
| 36 | 2017 | 2 | 6 | 00:00 | UTC | -1.0 | 7.0 | -7.3 |
| 37 | 2017 | 2 | 7 | 00:00 | UTC | -1.0 | 8.0 | -12.1 |
| 38 | 2017 | 2 | 8 | 00:00 | UTC | -1.0 | 8.0 | -8.8 |
| 39 | 2017 | 2 | 9 | 00:00 | UTC | -1.0 | 8.0 | -10.1 |
| 40 | 2017 | 2 | 10 | 00:00 | UTC | -1.0 | 8.0 | -8.3 |
| 41 | 2017 | 2 | 11 | 00:00 | UTC | -1.0 | 8.0 | -5.4 |
| 42 | 2017 | 2 | 12 | 00:00 | UTC | -1.0 | 8.0 | -2.7 |
| 43 | 2017 | 2 | 13 | 00:00 | UTC | -1.0 | 8.0 | 1.5 |
| 44 | 2017 | 2 | 14 | 00:00 | UTC | -1.0 | 8.0 | 4.4 |
| 45 | 2017 | 2 | 15 | 00:00 | UTC | -1.0 | 8.0 | 0.0 |
| 46 | 2017 | 2 | 16 | 00:00 | UTC | 0.9 | 8.0 | 0.5 |
| 47 | 2017 | 2 | 17 | 00:00 | UTC | 0.2 | 8.0 | 1.5 |
| 48 | 2017 | 2 | 18 | 00:00 | UTC | 1.5 | 5.0 | 1.9 |
| 49 | 2017 | 2 | 19 | 00:00 | UTC | 1.1 | 5.0 | 2.2 |
| 50 | 2017 | 2 | 20 | 00:00 | UTC | 2.8 | 3.0 | 0.4 |
| 51 | 2017 | 2 | 21 | 00:00 | UTC | -1.0 | 7.0 | -2.5 |
| 52 | 2017 | 2 | 22 | 00:00 | UTC | 12.2 | 6.0 | -4.6 |
| 53 | 2017 | 2 | 23 | 00:00 | UTC | 0.3 | 15.0 | -0.7 |
| 54 | 2017 | 2 | 24 | 00:00 | UTC | -1.0 | 13.0 | -5.3 |
| 55 | 2017 | 2 | 25 | 00:00 | UTC | 0.4 | 13.0 | -5.6 |
| 56 | 2017 | 2 | 26 | 00:00 | UTC | 2.5 | 12.0 | -2.0 |
| 57 | 2017 | 2 | 27 | 00:00 | UTC | 1.0 | 14.0 | -2.3 |
| 58 | 2017 | 2 | 28 | 00:00 | UTC | 7.7 | 13.0 | 2.1 |
The groupby object functions a bit like a DataFrame, so some operations which are allowed for DataFrames are also allowed for the groupby object. For example, we can get a subset of columns:
[32]:
groups["Temperature"]
[32]:
<pandas.core.groupby.groupby.SeriesGroupBy object at 0x7f97142b5f28>
For each DataFrame corresponding to a group the Temperature column was chosen. Still nothing was shown, because we haven’t applied any operation on the groups.
The common methods also include the aggregation methods. Let’s try to apply the mean aggregation:
[33]:
groups["Temperature"].mean()
[33]:
Month
1 -2.316129
2 -2.389286
3 0.983871
4 2.676667
5 9.783871
6 13.726667
7 16.035484
8 16.183871
9 11.826667
10 5.454839
11 3.950000
12 1.741935
Name: Temperature, dtype: float64
Now what happened was that after the mean aggregation was performed on each group, the results were automatically combined into a resulting DataFrame. Let’s try some other aggregation:
[34]:
groups["Precipitation"].sum()
[34]:
Month
1 26.9
2 21.0
3 29.7
4 26.9
5 -5.9
6 59.3
7 14.2
8 70.1
9 51.2
10 173.5
11 117.2
12 133.6
Name: Precipitation, dtype: float64
Ok, the -1.0 values in the Precipitation field are causing trouble here, let’s convert them to zeros:
[35]:
wh4 = wh3.copy()
wh4.loc[wh4.Precipitation == -1, "Precipitation"] = 0
wh4.loc[wh4.Snow == -1, "Snow"] = 0
wh4.head()
[35]:
| Year | Month | Day | Time | Time zone | Precipitation | Snow | Temperature | |
|---|---|---|---|---|---|---|---|---|
| 0 | 2017 | 1 | 1 | 00:00 | UTC | 0.0 | 0.0 | 0.6 |
| 1 | 2017 | 1 | 2 | 00:00 | UTC | 4.4 | 0.0 | -3.9 |
| 2 | 2017 | 1 | 3 | 00:00 | UTC | 6.6 | 7.0 | -6.5 |
| 3 | 2017 | 1 | 4 | 00:00 | UTC | 0.0 | 13.0 | -12.8 |
| 4 | 2017 | 1 | 5 | 00:00 | UTC | 0.0 | 10.0 | -17.8 |
[36]:
wh4.groupby("Month")["Precipitation"].sum()
[36]:
Month
1 38.9
2 35.0
3 41.7
4 39.9
5 16.1
6 76.3
7 31.2
8 86.1
9 65.2
10 184.5
11 120.2
12 140.6
Name: Precipitation, dtype: float64
Other ways to operate on groups¶
The aggregations are not the only possible operations on groups. The other possibilities are filtering, transformation, and application.
In filtering some of the groups can be filtered out.
[37]:
def myfilter(df): # The filter function must return a boolean value
return df["Precipitation"].sum() >= 150
wh4.groupby("Month").filter(myfilter) # Filter out months with total precipitation less that 150 mm
[37]:
| Year | Month | Day | Time | Time zone | Precipitation | Snow | Temperature | |
|---|---|---|---|---|---|---|---|---|
| 273 | 2017 | 10 | 1 | 00:00 | UTC | 0.0 | 0.0 | 9.1 |
| 274 | 2017 | 10 | 2 | 00:00 | UTC | 6.4 | 0.0 | 9.2 |
| 275 | 2017 | 10 | 3 | 00:00 | UTC | 21.5 | 0.0 | 8.3 |
| 276 | 2017 | 10 | 4 | 00:00 | UTC | 12.7 | 0.0 | 11.2 |
| 277 | 2017 | 10 | 5 | 00:00 | UTC | 0.6 | 0.0 | 8.8 |
| 278 | 2017 | 10 | 6 | 00:00 | UTC | 0.7 | 0.0 | 7.7 |
| 279 | 2017 | 10 | 7 | 00:00 | UTC | 11.7 | 0.0 | 8.1 |
| 280 | 2017 | 10 | 8 | 00:00 | UTC | 14.1 | 0.0 | 9.3 |
| 281 | 2017 | 10 | 9 | 00:00 | UTC | 18.3 | 0.0 | 8.6 |
| 282 | 2017 | 10 | 10 | 00:00 | UTC | 24.2 | 0.0 | 8.1 |
| 283 | 2017 | 10 | 11 | 00:00 | UTC | 1.5 | 0.0 | 6.9 |
| 284 | 2017 | 10 | 12 | 00:00 | UTC | 18.1 | 0.0 | 6.0 |
| 285 | 2017 | 10 | 13 | 00:00 | UTC | 0.0 | 0.0 | 7.5 |
| 286 | 2017 | 10 | 14 | 00:00 | UTC | 5.0 | 0.0 | 7.2 |
| 287 | 2017 | 10 | 15 | 00:00 | UTC | 3.3 | 0.0 | 8.3 |
| 288 | 2017 | 10 | 16 | 00:00 | UTC | 0.0 | 0.0 | 10.7 |
| 289 | 2017 | 10 | 17 | 00:00 | UTC | 1.6 | 0.0 | 8.5 |
| 290 | 2017 | 10 | 18 | 00:00 | UTC | 0.0 | 0.0 | 8.3 |
| 291 | 2017 | 10 | 19 | 00:00 | UTC | 0.9 | 0.0 | 4.6 |
| 292 | 2017 | 10 | 20 | 00:00 | UTC | 0.0 | 0.0 | 2.0 |
| 293 | 2017 | 10 | 21 | 00:00 | UTC | 0.0 | 0.0 | 0.2 |
| 294 | 2017 | 10 | 22 | 00:00 | UTC | 0.0 | 0.0 | 0.1 |
| 295 | 2017 | 10 | 23 | 00:00 | UTC | 0.0 | 0.0 | 1.3 |
| 296 | 2017 | 10 | 24 | 00:00 | UTC | 0.0 | 0.0 | 0.8 |
| 297 | 2017 | 10 | 25 | 00:00 | UTC | 8.5 | 0.0 | 2.1 |
| 298 | 2017 | 10 | 26 | 00:00 | UTC | 12.3 | 2.0 | 0.3 |
| 299 | 2017 | 10 | 27 | 00:00 | UTC | 2.7 | 7.0 | -0.3 |
| 300 | 2017 | 10 | 28 | 00:00 | UTC | 17.1 | 4.0 | 3.3 |
| 301 | 2017 | 10 | 29 | 00:00 | UTC | 3.3 | 0.0 | 2.1 |
| 302 | 2017 | 10 | 30 | 00:00 | UTC | 0.0 | 0.0 | 1.2 |
| 303 | 2017 | 10 | 31 | 00:00 | UTC | 0.0 | 0.0 | -0.4 |
In a transformation each group’s DataFrame is manipulated in a way that retains its shape. An example of centering values, so that the deviations from the monthly means are shown:
[38]:
pd.concat([wh4.iloc[:, 0:3],
wh4.groupby("Month")[["Precipitation", "Snow", "Temperature"]].transform(lambda x : x - x.mean())],
axis=1)
[38]:
| Year | Month | Day | Precipitation | Snow | Temperature | |
|---|---|---|---|---|---|---|
| 0 | 2017 | 1 | 1 | -1.254839 | -6.903226 | 2.916129 |
| 1 | 2017 | 1 | 2 | 3.145161 | -6.903226 | -1.583871 |
| 2 | 2017 | 1 | 3 | 5.345161 | 0.096774 | -4.183871 |
| 3 | 2017 | 1 | 4 | -1.254839 | 6.096774 | -10.483871 |
| 4 | 2017 | 1 | 5 | -1.254839 | 3.096774 | -15.483871 |
| 5 | 2017 | 1 | 6 | -0.954839 | 3.096774 | -15.483871 |
| 6 | 2017 | 1 | 7 | 4.045161 | 3.096774 | -1.483871 |
| 7 | 2017 | 1 | 8 | -1.254839 | 5.096774 | 1.816129 |
| 8 | 2017 | 1 | 9 | -0.154839 | 5.096774 | 2.816129 |
| 9 | 2017 | 1 | 10 | -0.954839 | 2.096774 | 4.016129 |
| 10 | 2017 | 1 | 11 | -1.254839 | 0.096774 | 0.716129 |
| 11 | 2017 | 1 | 12 | 6.745161 | 0.096774 | -0.483871 |
| 12 | 2017 | 1 | 13 | -1.154839 | 6.096774 | 3.416129 |
| 13 | 2017 | 1 | 14 | -1.154839 | 1.096774 | 3.116129 |
| 14 | 2017 | 1 | 15 | -1.254839 | 1.096774 | -0.483871 |
| 15 | 2017 | 1 | 16 | -1.254839 | 1.096774 | -1.883871 |
| 16 | 2017 | 1 | 17 | -1.054839 | 1.096774 | -1.183871 |
| 17 | 2017 | 1 | 18 | -0.354839 | 1.096774 | 3.416129 |
| 18 | 2017 | 1 | 19 | -1.254839 | -1.903226 | 3.916129 |
| 19 | 2017 | 1 | 20 | -0.954839 | -1.903226 | 1.716129 |
| 20 | 2017 | 1 | 21 | -0.854839 | -1.903226 | 0.516129 |
| 21 | 2017 | 1 | 22 | -1.054839 | -1.903226 | 3.316129 |
| 22 | 2017 | 1 | 23 | -1.154839 | -0.903226 | 2.416129 |
| 23 | 2017 | 1 | 24 | -1.254839 | -0.903226 | 0.116129 |
| 24 | 2017 | 1 | 25 | -0.654839 | -0.903226 | -1.483871 |
| 25 | 2017 | 1 | 26 | -1.254839 | -0.903226 | 4.216129 |
| 26 | 2017 | 1 | 27 | -1.254839 | -2.903226 | 3.916129 |
| 27 | 2017 | 1 | 28 | 0.545161 | -2.903226 | 3.116129 |
| 28 | 2017 | 1 | 29 | 1.345161 | -3.903226 | 2.916129 |
| 29 | 2017 | 1 | 30 | 4.345161 | -1.903226 | 3.316129 |
| ... | ... | ... | ... | ... | ... | ... |
| 335 | 2017 | 12 | 2 | 0.764516 | 3.516129 | -0.341935 |
| 336 | 2017 | 12 | 3 | 2.664516 | -1.483871 | 3.258065 |
| 337 | 2017 | 12 | 4 | -4.535484 | -1.483871 | -0.441935 |
| 338 | 2017 | 12 | 5 | -3.835484 | -1.483871 | -1.741935 |
| 339 | 2017 | 12 | 6 | -4.535484 | -1.483871 | -2.941935 |
| 340 | 2017 | 12 | 7 | 11.764516 | -1.483871 | -2.541935 |
| 341 | 2017 | 12 | 8 | -2.535484 | -1.483871 | 3.458065 |
| 342 | 2017 | 12 | 9 | -4.335484 | -1.483871 | 2.458065 |
| 343 | 2017 | 12 | 10 | -4.535484 | -1.483871 | 0.258065 |
| 344 | 2017 | 12 | 11 | -3.235484 | -1.483871 | -0.341935 |
| 345 | 2017 | 12 | 12 | 30.464516 | -1.483871 | -0.141935 |
| 346 | 2017 | 12 | 13 | -0.335484 | 3.516129 | -0.141935 |
| 347 | 2017 | 12 | 14 | 0.664516 | 2.516129 | -0.141935 |
| 348 | 2017 | 12 | 15 | 5.464516 | 8.516129 | -0.041935 |
| 349 | 2017 | 12 | 16 | -3.235484 | 4.516129 | 0.658065 |
| 350 | 2017 | 12 | 17 | -4.535484 | 3.516129 | -1.641935 |
| 351 | 2017 | 12 | 18 | -1.035484 | 3.516129 | 0.258065 |
| 352 | 2017 | 12 | 19 | -4.335484 | 1.516129 | -0.741935 |
| 353 | 2017 | 12 | 20 | -0.935484 | 1.516129 | 0.858065 |
| 354 | 2017 | 12 | 21 | -4.535484 | -1.483871 | 0.758065 |
| 355 | 2017 | 12 | 22 | -4.535484 | -1.483871 | -1.841935 |
| 356 | 2017 | 12 | 23 | 3.064516 | -1.483871 | -0.541935 |
| 357 | 2017 | 12 | 24 | -4.535484 | -1.483871 | -2.041935 |
| 358 | 2017 | 12 | 25 | 1.364516 | -1.483871 | -1.441935 |
| 359 | 2017 | 12 | 26 | 3.264516 | -1.483871 | 0.158065 |
| 360 | 2017 | 12 | 27 | -3.435484 | -1.483871 | 2.058065 |
| 361 | 2017 | 12 | 28 | -0.835484 | -1.483871 | 1.058065 |
| 362 | 2017 | 12 | 29 | 3.264516 | -1.483871 | 2.058065 |
| 363 | 2017 | 12 | 30 | -0.435484 | -1.483871 | 0.758065 |
| 364 | 2017 | 12 | 31 | -1.335484 | -1.483871 | -0.141935 |
365 rows × 6 columns
The apply method is very generic and only requires that for each group’s DataFrame the given function returns a DataFrame, Series, or a scalar. In the following example, we sort within each group by the temperature:
[39]:
wh4.groupby("Month").apply(lambda df : df.sort_values("Temperature"))
[39]:
| Year | Month | Day | Time | Time zone | Precipitation | Snow | Temperature | ||
|---|---|---|---|---|---|---|---|---|---|
| Month | |||||||||
| 1 | 4 | 2017 | 1 | 5 | 00:00 | UTC | 0.0 | 10.0 | -17.8 |
| 5 | 2017 | 1 | 6 | 00:00 | UTC | 0.3 | 10.0 | -17.8 | |
| 3 | 2017 | 1 | 4 | 00:00 | UTC | 0.0 | 13.0 | -12.8 | |
| 2 | 2017 | 1 | 3 | 00:00 | UTC | 6.6 | 7.0 | -6.5 | |
| 15 | 2017 | 1 | 16 | 00:00 | UTC | 0.0 | 8.0 | -4.2 | |
| 1 | 2017 | 1 | 2 | 00:00 | UTC | 4.4 | 0.0 | -3.9 | |
| 24 | 2017 | 1 | 25 | 00:00 | UTC | 0.6 | 6.0 | -3.8 | |
| 6 | 2017 | 1 | 7 | 00:00 | UTC | 5.3 | 10.0 | -3.8 | |
| 16 | 2017 | 1 | 17 | 00:00 | UTC | 0.2 | 8.0 | -3.5 | |
| 11 | 2017 | 1 | 12 | 00:00 | UTC | 8.0 | 7.0 | -2.8 | |
| 14 | 2017 | 1 | 15 | 00:00 | UTC | 0.0 | 8.0 | -2.8 | |
| 23 | 2017 | 1 | 24 | 00:00 | UTC | 0.0 | 6.0 | -2.2 | |
| 20 | 2017 | 1 | 21 | 00:00 | UTC | 0.4 | 5.0 | -1.8 | |
| 10 | 2017 | 1 | 11 | 00:00 | UTC | 0.0 | 7.0 | -1.6 | |
| 19 | 2017 | 1 | 20 | 00:00 | UTC | 0.3 | 5.0 | -0.6 | |
| 7 | 2017 | 1 | 8 | 00:00 | UTC | 0.0 | 12.0 | -0.5 | |
| 22 | 2017 | 1 | 23 | 00:00 | UTC | 0.1 | 6.0 | 0.1 | |
| 30 | 2017 | 1 | 31 | 00:00 | UTC | 0.0 | 4.0 | 0.2 | |
| 8 | 2017 | 1 | 9 | 00:00 | UTC | 1.1 | 12.0 | 0.5 | |
| 28 | 2017 | 1 | 29 | 00:00 | UTC | 2.6 | 3.0 | 0.6 | |
| 0 | 2017 | 1 | 1 | 00:00 | UTC | 0.0 | 0.0 | 0.6 | |
| 13 | 2017 | 1 | 14 | 00:00 | UTC | 0.1 | 8.0 | 0.8 | |
| 27 | 2017 | 1 | 28 | 00:00 | UTC | 1.8 | 4.0 | 0.8 | |
| 29 | 2017 | 1 | 30 | 00:00 | UTC | 5.6 | 5.0 | 1.0 | |
| 21 | 2017 | 1 | 22 | 00:00 | UTC | 0.2 | 5.0 | 1.0 | |
| 12 | 2017 | 1 | 13 | 00:00 | UTC | 0.1 | 13.0 | 1.1 | |
| 17 | 2017 | 1 | 18 | 00:00 | UTC | 0.9 | 8.0 | 1.1 | |
| 18 | 2017 | 1 | 19 | 00:00 | UTC | 0.0 | 5.0 | 1.6 | |
| 26 | 2017 | 1 | 27 | 00:00 | UTC | 0.0 | 4.0 | 1.6 | |
| 9 | 2017 | 1 | 10 | 00:00 | UTC | 0.3 | 9.0 | 1.7 | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 12 | 340 | 2017 | 12 | 7 | 00:00 | UTC | 16.3 | 0.0 | -0.8 |
| 357 | 2017 | 12 | 24 | 00:00 | UTC | 0.0 | 0.0 | -0.3 | |
| 355 | 2017 | 12 | 22 | 00:00 | UTC | 0.0 | 0.0 | -0.1 | |
| 338 | 2017 | 12 | 5 | 00:00 | UTC | 0.7 | 0.0 | 0.0 | |
| 350 | 2017 | 12 | 17 | 00:00 | UTC | 0.0 | 5.0 | 0.1 | |
| 358 | 2017 | 12 | 25 | 00:00 | UTC | 5.9 | 0.0 | 0.3 | |
| 334 | 2017 | 12 | 1 | 00:00 | UTC | 3.4 | 0.0 | 0.9 | |
| 352 | 2017 | 12 | 19 | 00:00 | UTC | 0.2 | 3.0 | 1.0 | |
| 356 | 2017 | 12 | 23 | 00:00 | UTC | 7.6 | 0.0 | 1.2 | |
| 337 | 2017 | 12 | 4 | 00:00 | UTC | 0.0 | 0.0 | 1.3 | |
| 335 | 2017 | 12 | 2 | 00:00 | UTC | 5.3 | 5.0 | 1.4 | |
| 344 | 2017 | 12 | 11 | 00:00 | UTC | 1.3 | 0.0 | 1.4 | |
| 364 | 2017 | 12 | 31 | 00:00 | UTC | 3.2 | 0.0 | 1.6 | |
| 346 | 2017 | 12 | 13 | 00:00 | UTC | 4.2 | 5.0 | 1.6 | |
| 345 | 2017 | 12 | 12 | 00:00 | UTC | 35.0 | 0.0 | 1.6 | |
| 347 | 2017 | 12 | 14 | 00:00 | UTC | 5.2 | 4.0 | 1.6 | |
| 348 | 2017 | 12 | 15 | 00:00 | UTC | 10.0 | 10.0 | 1.7 | |
| 359 | 2017 | 12 | 26 | 00:00 | UTC | 7.8 | 0.0 | 1.9 | |
| 351 | 2017 | 12 | 18 | 00:00 | UTC | 3.5 | 5.0 | 2.0 | |
| 343 | 2017 | 12 | 10 | 00:00 | UTC | 0.0 | 0.0 | 2.0 | |
| 349 | 2017 | 12 | 16 | 00:00 | UTC | 1.3 | 6.0 | 2.4 | |
| 363 | 2017 | 12 | 30 | 00:00 | UTC | 4.1 | 0.0 | 2.5 | |
| 354 | 2017 | 12 | 21 | 00:00 | UTC | 0.0 | 0.0 | 2.5 | |
| 353 | 2017 | 12 | 20 | 00:00 | UTC | 3.6 | 3.0 | 2.6 | |
| 361 | 2017 | 12 | 28 | 00:00 | UTC | 3.7 | 0.0 | 2.8 | |
| 360 | 2017 | 12 | 27 | 00:00 | UTC | 1.1 | 0.0 | 3.8 | |
| 362 | 2017 | 12 | 29 | 00:00 | UTC | 7.8 | 0.0 | 3.8 | |
| 342 | 2017 | 12 | 9 | 00:00 | UTC | 0.2 | 0.0 | 4.2 | |
| 336 | 2017 | 12 | 3 | 00:00 | UTC | 7.2 | 0.0 | 5.0 | |
| 341 | 2017 | 12 | 8 | 00:00 | UTC | 2.0 | 0.0 | 5.2 |
365 rows × 8 columns
This exercise can give two points at maximum!
Part 1.
Read, clean and parse the bicycle data set as before. Group the rows by year, month, and day. Get the sum for each group. Make function cyclists_per_day that does the above. The function should return a DataFrame. Make sure that the columns Hour and Weekday are not included in the returned DataFrame.
Part 2.
In the main function, using the function cyclists_per_day, get the daily counts. The index of the DataFrame now consists of tuples (Year, Month, Day). Then restrict this data to August of year 2017, and plot this data. Don’t forget to call the plt.show function of matplotlib. The x-axis should have ticks from 1 to 31, and there should be a curve to each measuring station. Can you spot the weekends?
We use again the UK top 40 data set from the first week of 1964 in the src folder. Here we define “goodness” of a record company (Publisher) based on the sum of the weeks on chart (WoC) of its singles. Return a DataFrame of the singles by the best record company (a subset of rows of the original DataFrame). Do this with function best_record_company.
Load the suicide data set from src folder. This data was originally downloaded from Kaggle. Kaggle contains lots of interesting open data sets.
Write function suicide_fractions that loads the data set and returns a Series that has the country as the (row) index and as the column the mean fraction of suicides per population in that country. In other words, the value is the average of suicide fractions. The information about year, sex and age is not used.
Copy the function suicide fractions from the previous exercise.
Implement function suicide_weather as described below. We use the dataset of average temperature (over years 1961-1990) in different countries from src/List_of_countries_by_average_yearly_temperature.html (https://en.wikipedia.org/wiki/List_of_countries_by_average_yearly_temperature) . You can use the function pd.read_html to get all the tables from a html page. By default pd.read_html does not know which row contains column headers and which column contains row headers.
Therefore, you have to give both index_col and header parameters to read_html. Maku sure you use the country as the (row) index for both of the DataFrames. What is the Spearman correlation between these variables? Use the corr method of Series object. Note the the two Series need not be sorted as the indices of the rows (country names) are used to align them.
The return value of the function suicide_weather is a tuple (suicide_rows, temperature_rows, common_rows, spearman_correlation) The output from the main function should be of the following form:
Suicide DataFrame has x rows
Temperature DataFrame has x rows
Common DataFrame has x rows
Spearman correlation: x.x
You might have trouble when trying to convert the temperatures to float. The is because the negative numbers on that html page use a special unicode minus sign, which looks typographically nice, but the float constructor cannot interpret it as a minus sign. You can try out the following example:
[40]:
s="\u2212" "5" # unicode minus sign and five
print(s)
try:
float(s)
except ValueError as e:
import sys
print(e, file=sys.stderr)
−5
could not convert string to float: '−5'
But if we explicitly convert unicode minus sign to normal minus sign, it works:
[41]:
float(s.replace("\u2212", "-"))
[41]:
-5.0
Time series¶
If a measurement is made at certain points in time, the resulting values with their measurement times is called a time series. In Pandas a Series whose index consists of dates/times is a time series.
Let’s make a copy of the DataFrame that we can mess with:
[42]:
wh2 = wh3.copy()
wh2.columns
[42]:
Index(['Year', 'Month', 'Day', 'Time', 'Time zone', 'Precipitation', 'Snow',
'Temperature'],
dtype='object')
The column names Year, Month, and Day are now in appropriate form for the to_datetime function. It can convert these fields into a timestamp series, which we will add to the DataFrame.
[43]:
wh2["Date"] = pd.to_datetime(wh2[["Year", "Month", "Day"]])
wh2.head()
[43]:
| Year | Month | Day | Time | Time zone | Precipitation | Snow | Temperature | Date | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 2017 | 1 | 1 | 00:00 | UTC | -1.0 | -1.0 | 0.6 | 2017-01-01 |
| 1 | 2017 | 1 | 2 | 00:00 | UTC | 4.4 | -1.0 | -3.9 | 2017-01-02 |
| 2 | 2017 | 1 | 3 | 00:00 | UTC | 6.6 | 7.0 | -6.5 | 2017-01-03 |
| 3 | 2017 | 1 | 4 | 00:00 | UTC | -1.0 | 13.0 | -12.8 | 2017-01-04 |
| 4 | 2017 | 1 | 5 | 00:00 | UTC | -1.0 | 10.0 | -17.8 | 2017-01-05 |
We can now drop the useless fields:
[44]:
wh2=wh2.drop(columns=["Year", "Month", "Day"])
wh2.head()
[44]:
| Time | Time zone | Precipitation | Snow | Temperature | Date | |
|---|---|---|---|---|---|---|
| 0 | 00:00 | UTC | -1.0 | -1.0 | 0.6 | 2017-01-01 |
| 1 | 00:00 | UTC | 4.4 | -1.0 | -3.9 | 2017-01-02 |
| 2 | 00:00 | UTC | 6.6 | 7.0 | -6.5 | 2017-01-03 |
| 3 | 00:00 | UTC | -1.0 | 13.0 | -12.8 | 2017-01-04 |
| 4 | 00:00 | UTC | -1.0 | 10.0 | -17.8 | 2017-01-05 |
The following method call will set the Date field as the index of the DataFrame.
[45]:
wh2 = wh2.set_index("Date")
wh2.head()
[45]:
| Time | Time zone | Precipitation | Snow | Temperature | |
|---|---|---|---|---|---|
| Date | |||||
| 2017-01-01 | 00:00 | UTC | -1.0 | -1.0 | 0.6 |
| 2017-01-02 | 00:00 | UTC | 4.4 | -1.0 | -3.9 |
| 2017-01-03 | 00:00 | UTC | 6.6 | 7.0 | -6.5 |
| 2017-01-04 | 00:00 | UTC | -1.0 | 13.0 | -12.8 |
| 2017-01-05 | 00:00 | UTC | -1.0 | 10.0 | -17.8 |
We can now easily get a set of rows using date slices:
[46]:
wh2["2017-01-15":"2017-02-03"]
[46]:
| Time | Time zone | Precipitation | Snow | Temperature | |
|---|---|---|---|---|---|
| Date | |||||
| 2017-01-15 | 00:00 | UTC | -1.0 | 8.0 | -2.8 |
| 2017-01-16 | 00:00 | UTC | -1.0 | 8.0 | -4.2 |
| 2017-01-17 | 00:00 | UTC | 0.2 | 8.0 | -3.5 |
| 2017-01-18 | 00:00 | UTC | 0.9 | 8.0 | 1.1 |
| 2017-01-19 | 00:00 | UTC | -1.0 | 5.0 | 1.6 |
| 2017-01-20 | 00:00 | UTC | 0.3 | 5.0 | -0.6 |
| 2017-01-21 | 00:00 | UTC | 0.4 | 5.0 | -1.8 |
| 2017-01-22 | 00:00 | UTC | 0.2 | 5.0 | 1.0 |
| 2017-01-23 | 00:00 | UTC | 0.1 | 6.0 | 0.1 |
| 2017-01-24 | 00:00 | UTC | -1.0 | 6.0 | -2.2 |
| 2017-01-25 | 00:00 | UTC | 0.6 | 6.0 | -3.8 |
| 2017-01-26 | 00:00 | UTC | -1.0 | 6.0 | 1.9 |
| 2017-01-27 | 00:00 | UTC | -1.0 | 4.0 | 1.6 |
| 2017-01-28 | 00:00 | UTC | 1.8 | 4.0 | 0.8 |
| 2017-01-29 | 00:00 | UTC | 2.6 | 3.0 | 0.6 |
| 2017-01-30 | 00:00 | UTC | 5.6 | 5.0 | 1.0 |
| 2017-01-31 | 00:00 | UTC | -1.0 | 4.0 | 0.2 |
| 2017-02-01 | 00:00 | UTC | 1.5 | 4.0 | -0.6 |
| 2017-02-02 | 00:00 | UTC | 0.2 | 5.0 | -0.8 |
| 2017-02-03 | 00:00 | UTC | -1.0 | 6.0 | -0.2 |
By using the date_range function even more complicated sets can be formed. The following gets all the Mondays of July:
[47]:
r=pd.date_range("2017-07-01", "2017-07-31", freq="w-mon")
r
[47]:
DatetimeIndex(['2017-07-03', '2017-07-10', '2017-07-17', '2017-07-24',
'2017-07-31'],
dtype='datetime64[ns]', freq='W-MON')
[48]:
wh2.index.difference(r)
[48]:
DatetimeIndex(['2017-01-01', '2017-01-02', '2017-01-03', '2017-01-04',
'2017-01-05', '2017-01-06', '2017-01-07', '2017-01-08',
'2017-01-09', '2017-01-10',
...
'2017-12-22', '2017-12-23', '2017-12-24', '2017-12-25',
'2017-12-26', '2017-12-27', '2017-12-28', '2017-12-29',
'2017-12-30', '2017-12-31'],
dtype='datetime64[ns]', length=360, freq=None)
[49]:
wh2.loc[r,:]
[49]:
| Time | Time zone | Precipitation | Snow | Temperature | |
|---|---|---|---|---|---|
| 2017-07-03 | 00:00 | UTC | 2.2 | -1.0 | 14.5 |
| 2017-07-10 | 00:00 | UTC | -1.0 | -1.0 | 18.0 |
| 2017-07-17 | 00:00 | UTC | 2.7 | -1.0 | 15.4 |
| 2017-07-24 | 00:00 | UTC | -1.0 | -1.0 | 15.7 |
| 2017-07-31 | 00:00 | UTC | 0.1 | -1.0 | 17.8 |
The following finds all the business days (Monday to Friday) of July:
[50]:
pd.date_range("2017-07-01", "2017-07-31", freq="b")
[50]:
DatetimeIndex(['2017-07-03', '2017-07-04', '2017-07-05', '2017-07-06',
'2017-07-07', '2017-07-10', '2017-07-11', '2017-07-12',
'2017-07-13', '2017-07-14', '2017-07-17', '2017-07-18',
'2017-07-19', '2017-07-20', '2017-07-21', '2017-07-24',
'2017-07-25', '2017-07-26', '2017-07-27', '2017-07-28',
'2017-07-31'],
dtype='datetime64[ns]', freq='B')
We can get a general idea about the Temperature column by plotting it. Note how the index time series is shown nicely on the x-axis.
[51]:
%matplotlib inline
wh2["Temperature"].plot();
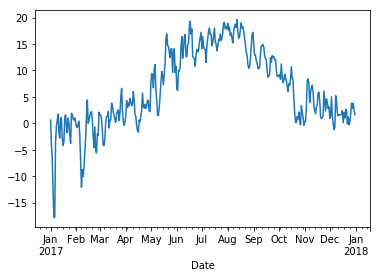
The graph looks a bit messy at this level of detail. We can smooth it by taking averages over a sliding window of length 30 days:
[52]:
rolling = wh2.Temperature.rolling(30, center=True)
rolling
[52]:
Rolling [window=30,center=True,axis=0]
[53]:
data = pd.DataFrame({"Temperature" : wh2.Temperature, "Rolling mean" : rolling.mean()})
data.plot();
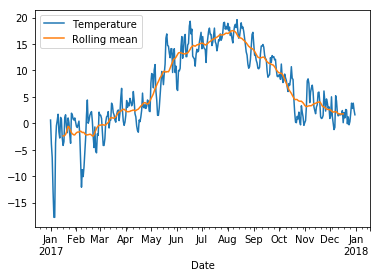
Write function bicycle_timeseries that
- reads the data set
- cleans it
- turns its
Päivämääräcolumn into (row) DatetimeIndex (that is, to row names) of that DataFrame - returns the new DataFrame
In function commute do the following:
Use the function bicycle_timeseries to get the bicycle data. Restrict to August 2017, group by the weekday, aggregate by summing. Set the Weekday column to numbers from one to seven. Then set the column Weekday as the (row) index. Return the resulting DataFrame from the function.
In the main function plot the DataFrame. Xticklabels should be the weekdays. Don’t forget to call show function!
If you want the xticklabels to be ['Mon', 'Tue', 'Wed', 'Thu', 'Fr', 'Sat', 'Sun'] instead of numbers (1,..,7), then it may get a bit messy. There seems to be a problem with non-numeric x values. You could try the following after plotting, but you don’t have to:
weekdays="x mon tue wed thu fri sat sun".title().split()
plt.gca().set_xticklabels(weekdays)
Additional information¶
Pandas cheat sheet Summary of most important Pandas’ functions and methods.
Read the article Tidy Data. The article uses the statistical software R as an example, but the ideas are relevant in general. Pandas operations maintain data in the tidy format.
Pandas handles only one dimensional data (Series) and two dimensional data (DataFrame). While you can use hierarchical indices to simulate higher dimensional arrays, you should use the xarray library, if you need proper higher-dimensional arrays with labels. It is basically a cross between NumPy and Pandas.